枝角类是淡水生态系统中浮游动物的重要类群,也是水体食物网的主要组分.枝角类通常具有两种生殖方式:一般情况下,枝角类以孤雌生殖为主,此时雌体所产的卵称为孤雌生殖卵,因不需要受精,也称非受精卵;在环境条件发生不利的变化(极端温度、干旱、缺氧等)时,一些孤雌生殖卵会发育成雄体,与双性雌体交配,进行有性生殖,产生的有性生殖卵称为休眠卵.休眠卵在母体脱壳时,与壳瓣一同脱出,受水流及风浪的影响漂浮至沿岸带或者沉入水底[1].不同时期产生的具有萌发活性的休眠卵沉积到水体底部,以休眠卵的形式形成种子库[2].休眠卵库导致了世代周期重叠,形成了种群的存储机制,这种机制有利于维护种群遗传多样性[3-4],并在不利环境条件下,使种群在时间尺度上得到延续[5-7].
对一个具体的水体而言,我们通常是以枝角类现生种群为对象,通过测定现生种群的遗传信息来推断其遗传结构.对产生休眠卵的枝角类而言,由现生种群产生的这些可完好保存于底泥中的休眠卵形成了不同世代现生种群的种子库,特别是位于底泥表层2~10 cm中的休眠卵具有较高的活性,容易获得环境条件的直接诱导萌发,休眠卵萌发后形成的个体对于现生种群不仅是数量上的补充,同时也进行了遗传上的补充,这种遗传补充也可称为水体内部发生的跨世代的基因流(持续奠基者效应)[8],在很大程度上影响枝角类现生种群的遗传多样性和结构.然而,枝角类在通常情况下依靠孤雌生殖来快速扩张其种群数量,如果在缺少来自外部和休眠卵库的持续基因流时,孤雌生殖产生的不同克隆系,在强选择压力下被排除在水体之外,导致水体现生种群的遗传多样性下降.在时间尺度上,这一过程被称为种群遗传多样性的克隆侵蚀[9],这种现象在一些轮虫和大型枝角类的研究中得到证实[10-13].
在温带水体,枝角类生长季节初期,卵库中的枝角类休眠卵孵化,进入现生种群,形成休眠种群到现生种群的基因流,增加现生种群遗传多样性,经过整个生长季节的克隆侵蚀,现生种群遗传多样性下降并通过有性生殖产生的休眠卵进入休眠卵库,形成现生种群到休眠种群的基因流,因此,休眠种群与现生种群存在一个双向的基因流过程[9].在热带亚热带水库中,由于全年的水温较高,有利于枝角类在水体中常年存在,但诱导枝角类形成休眠卵的外部条件不如温带水体强烈,休眠卵库在规模上(保存的种类少、数量低)也大大小于温带水体[14].另一方面,热带和亚热带水库的枝角类面临鱼类的选择压力要强于温带地区,在夏季鱼类捕食压力达到最大时,大型枝角类种群数量降至最低时(常规采样难以获得)[15-16],大型枝角类以小种群通过垂直迁移的方式逃避鱼类捕食[16].因此,在热带亚热带大型深水水库中,根据大型枝角类休眠卵库对现生种群的补充相对较弱[16],即休眠种群到现生种群的基因流较弱,同时,鱼类捕食压力较强,现生种群面临的克隆侵蚀较强,我们推断现生种群与休眠种群的遗传结构和遗传多样性会存在较大差异.本文以热带水库——广东流溪河水库大型枝角类盔型溞(Daphnia galeata)为研究对象,利用线粒体DNA信息,比较分析盔形溞现生种群和休眠种群的遗传结构和遗传多样性是否存在较大差异.
1 材料与方法 1.1 样本采集与DNA提取于2010年5月,在广东省流溪河水库枝角类丰度较高的敞水区,同时考虑沉积特点选择采样点(23°45′N,113°46′E),使用Uwitec柱状采泥器(奥地利)采集沉积物.分别采集4~5柱保存较好的沉积物,沉积物长度为30~40 cm.每柱沉积物取表层10 cm现场切割后转移至封口袋中,为避免由于采泥器PVC柱子滑动造成不同泥层间的沉积物交叉污染,泥层外围的沉积物切割后需要舍弃[17-18].切割后的样品分别用孔径为600、200和35 μm的网筛依次过滤筛选,过滤后残留的液体转移至100 ml塑料小白瓶中,保存于4℃冰箱中.分离出含有休眠卵的残余液体在20~50倍放大的解剖镜(Olympus SZX71)和显微镜(Olympus CX21)下进行观察,挑取盔型溞休眠卵[19],并转移至无水乙醇中于4℃下保存.现生种群使用110 μm浮游生物网定性拖网采集,现场用75 %乙醇固定.带回实验室,镜检后置换无水乙醇,并置于4℃下保存. DNA的提取参考Montero-Pau等[20]和徐磊等[21]的方法,使用试剂盒提取溞属DNA.
1.2 线粒体COI序列扩增与测序COI基因扩增引物为ZplankF1(TGTAAAACGACGGCCAGTTCTASWAATCATAARGATATTGG)和ZplankR1(CAGGAAACAGCTATGACTTCAGGRTGRCCRAARAATCA)[22].扩增反应总体积为30 μl,包括:3 μl 10×buffer(Mg2+plus),dNTP Mixture 2.5 mmol/L,引物各0.5 μmol/L,0.5 U Taq酶(TaKaRa)和3 μl模板DNA.扩增条件为95℃预变性1 min,94℃变性40 s,51℃退火40 s,72℃延伸1 min,共35个循环;72℃ 3 min充分延伸4℃结束.扩增产物经凝胶电泳检测后,选取目的条带清晰且亮度大的PCR反应产物送华诺基因进行测序.所有样品均采用双向测序,正反向互补拼接后得到较完整序列.现生种群和休眠种群提取DNA个体数为20~30个个体或休眠卵.
1.3 数据处理及系统学分析 1.3.1 数据处理测序获得的COI基因序列先在NCBI数据库中进行BLAST搜索比对以保证序列的可靠性.用BioEdit 7[23]软件及其附带的Clustal W比对软件将实验获得的和GenBanK中下载的COI基因序列进行同源比对,获得的COI基因序列用于分析.单倍型多样性及核算多样性等在DnaSP 5.10软件[24]中分析计算.遗传差异度在MEGA 6.06软件[25]中分析,其中COI基因序列遗传差异度采用Kimura双参模型(Kimura 2-parameter)计算;变异估算方法为自举法(bootstrapping method),运行次数为1000;核苷酸替代包括转换和颠换,序列间位点选择相同进化率,删除缺失位点.在HAPLOVIEWER中构建单倍型网络图[26].根据溞属枝角类线粒体基因的突变率[27],使用Migrate-n version 3.6软件计算有效群体大小(Ne)及群体间的长期有效迁移率M(M=m/μ,m为每世代有效种群迁移率,μ为供点突变率),判断种群间基因流强度分析使用Bayesian MCMC方法(迭代次数为10000,重复计算5次,加热方案为默认的1.0、1.5、3、1000000).有效种群大小是指在一个理想种群中,和该种群随机遗传漂变下的等位基因传播或者近亲繁殖等同的个体数量.
1.3.2 系统学分析构建COI系统树之前,将获得的所有序列用Mrmodeltest 2.3软件进行AIC最适核酸替换模型筛选[28],选用MrBayes v. 3.1.2[29-30]构建贝叶斯系统树.详细参数设置如下:同时起始4条马尔科夫链,即3条热链和l条冷链,以随机树为起始树,共运行1000000代,每100代抽样1次,重复1次.在舍弃老化样本后,根据剩余的样本构建一致树.以后验概率来表示各分支的置信度,95 %以上为接受PP值,认定为可靠分支.外类群选自长刺溞复合种(Daphnia longispina complex)中的Daphnia dentifera和Daphnia longispina,序列来自NCBI(登录号为KM555366;EF375860).分析完成的系统树通过treeviewv. 1.6.6[31]展示.
2 结果 2.1 COI基因分歧度和单倍型共获得71条COI序列(每条COI基因序列对应1个个体),长度为581 bp,其中初期现生种群24个,末期现生种群20个,休眠种群27个(表 1). COI基因序列比对结果显示序列可识别位点(包括缺失与丢失的位点)581个,其中保守位点546个,变异位点35个,单一位点23个,简约信息点12个.基于Kimura双参数模型计算的初期现生种群遗传差异度为0.1 %,末期现生种群遗传差异度为0.33 %,休眠种群的遗传差异度为0.77 %,初期和末期现生种群与休眠种群间的遗传差异度均为0.6 %,初期现生种群与末期现生种群间的遗传差异度为0.2 % (表 2).初期现生种群、末期现生种群和休眠种群的单倍型多样性分别为0.562、0.726和0.815:核酸多样性分别为0.00104、0.00331和0.00761(表 1).现生种群获得的44条COI基因序列和休眠种群获得的27条COI基因序列共产生15个单倍型(图 1).
| 表 1 流溪河水库盔型溞现生种群和休眠种群的单倍型多样性和核酸多样性 Tab.1 The haplotype diversity and nucleotide diversity between dormant and active populations of Daphnia galeata in Liuxihe Reservoir |
| 表 2 流溪河水库盔形溞现生种群和休眠种群种间和种内的遗传差异度(%) Tab.2 The genetic differentiation between dormant and active populations of Daphnia galeata in Liuxihe Reservoir |

|
图 1 基于COI基因序列构建的盔型溞现生种群和休眠种群的单倍型网络 Fig.1 Haplotype network for the dormant and active populations of Daphnia galeata with mitochondria COI gene sequences |
通过Migrate-n version 3.6软件基于溯祖理论对种群间基因流水平进行分析,并使用群体间的长期有效迁移率M对基因流强度进行描述. M值衡量的是在一定突变率下,每个世代迁入的个体对整个种群遗传变异的影响程度.分析发现休眠种群的有效种群为192107,休眠种群到现生种群的M值为527.5,现生种群的有效种群为7285,现生种群到休眠种群的M值为490.9,流溪河盔型溞现生种群与休眠种群存在双向较强的基因流.
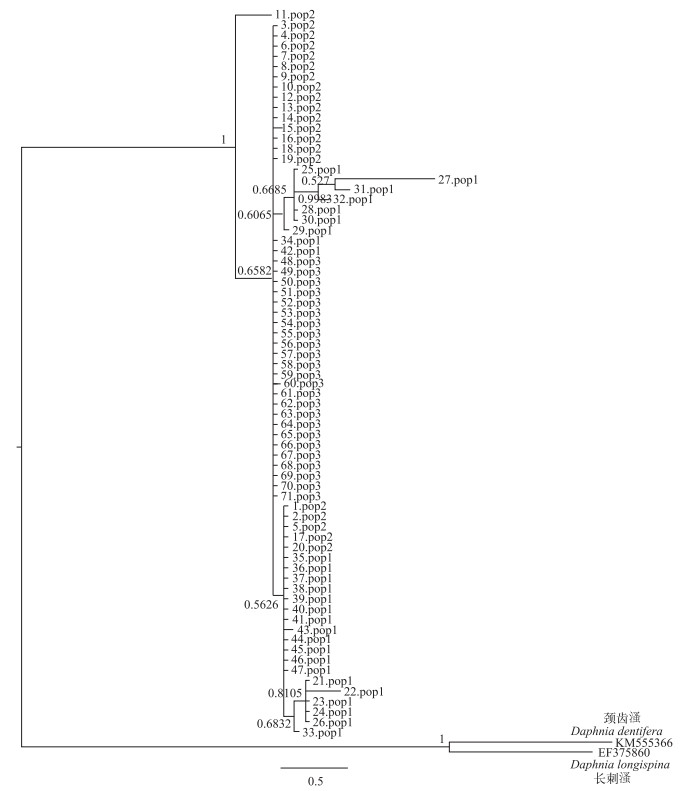
2.3 系统学分析由贝叶斯系统进化树(图 2)可知,休眠种群与现生种群间并未出现较大分化,且并不存在隐种或者亚种的分化.
|
图 2 基于COI基因序列构建盔形溞现生种群和休眠种群贝叶斯系统进化树(pop1为休眠种群;pop2为末期现生种群;pop3为初期现生种群) Fig.2 Bayes tree for the dormant and active populations of Daphnia galeata with mitochondria COI gene sequences(pop1 is dormant population; pop2 is end active population; pop3 is initial active population) |
流溪河盔型溞的休眠种群相比现生种群有着较高的单倍型多样性(克隆多样性)(图 1和表 1),这与休眠卵库的形成过程和补充机制有关.当一个水体新形成时,不同水体的盔型溞孤雌生殖个体或者休眠卵通过水流、鸟类迁徙、人类活动等途径进入水体,通过快速的孤雌生殖方式,快速占据水体中的生态位,形成稳定种群(克隆系组成),直到生长季节末期,水温或其他水环境条件发生改变,盔型溞通过有性生殖产生休眠卵,进入休眠卵库.春季枝角类生长季节初期,底泥中的休眠卵随着水体全同温水流扰动等影响,迁移至水层区或沿岸带萌发诱导,萌发后的盔型溞通过孤雌生殖形成现生种群.最终,休眠种群与现生种群形成一个动态平衡过程,而随后外源新迁入的盔型溞(新的克隆系)面临现生种群的竞争优势与该生境下的选择压力,难以直接形成有效的基因流进入现生种群[32],更多是以休眠卵的形式保存在底泥中,等待获得萌发的机会,经过长期的积累,使得休眠种群具有较高的遗传多样性(克隆多样性).
与休眠种群不同,现生种群在生长季节初期完全依赖于休眠种群的补充,即休眠种群的萌发,决定了生长季节初期现生种群的遗传结构.流溪河水库处于在我国热带亚热带地区,枝角类群落结构以及所面临的选择压力与温带湖泊都有着明显的区别[15-16].由于全年的水温较高,有利于一些枝角类种类的种群在水体中常年存在,诱导枝角类形成休眠卵的外部条件(如低温)不如温带水体强烈,导致热带水体休眠卵库相对较小[14].此外,我国热带亚热带地区的水库受人为控制水量(防洪、发电和灌溉等)的影响,休眠卵库的动态变化与水库的水文过程有着更为紧密的联系.特别是受季风气候影响降水较为集中的雨季,水库水位波动和水滞留时间变化较大,沿岸带形成的休眠卵被水流冲刷带入敞水区,因此而减小接触孵化刺激的机会.而在敞水区形成的休眠卵却很少有机会进入浅水区,现生种群和休眠种群的种群数量都会受水流冲刷带来的平流损失的影响,从而使得现生种群缺失来自于休眠卵库的持续补充[33-34].另一方面,热带亚热带水库中,鱼类捕食压力较强,枝角类生长季节较短[15],枝角类现生种群的遗传多样性面临的克隆侵蚀较强.而本文所采集的现生种群为生长季节初期和末期种群,在经历了整个生长季节的克隆侵蚀后[9, 35],现生种群的单倍型多样性降至最低,因此,流溪河盔型溞的休眠种群相比现生种群有着较高的单倍型多样性(克隆多样性).
3.2 休眠种群与现生种群的基因流我们假设在热带亚热带大型深水水库中,由于大型枝角类休眠卵数量较小(密度为103~104 ind./m2),比温度地区水体低1~2个数量级[14],休眠种群对现生种群的补充相对较弱,即休眠种群到现生种群的基因流较弱,然而,研究结果显示,流溪河盔型溞现生种群与休眠种群存在双向较强的基因流,现生种群到休眠种群的M值为490.9,休眠种群到现生种群的M值为527.5,这与我们的推断不同,由此可见,休眠种群到现生种群的基因流与休眠卵库大小无关,但与休眠卵的萌发有关.不同时期的现生种群单倍型多样性对比发现,生长季节初期现生种群的单倍型多样性低于生长季节末期,可见,整个生长季节过程中,休眠卵不断从休眠卵库萌发,补充进入现生种群.温带地区的研究表明,溞属枝角类休眠卵的萌发主要与水温有关,孵化时间与水温呈负相关.以蚤状溞(Daphnia pulex)为例,当水温为7℃时,孵化时间在10 d以上,当水温达到12℃时,孵化时间为7~10 d,而当水温达到15℃时,孵化时间为3~6 d,对于大型溞(Daphnia magna),前7 d的萌发率达到66 %,15 d的萌发率更是高达81.9 %,由休眠卵孵化的个体中,至少有57 %的个体可以发育为成体,完成一次孤雌生殖[36-40].如此高的休眠卵萌发效率,使得在温带及高海拔分布的溞属枝角类休眠种群可以在湖泊冰层消失之后的两周内迅速萌发,进入现生种群[41].在流溪河水库,冬季(12-2月)底层平均水温为13℃,而在枝角类生长季节初期(2月末,3月初)底层平均水温可达15℃以上,意味着盔型溞的休眠卵可以达到最适萌发条件,并在短时间内完成萌发,因此,较高的萌发率和较短的萌发时间使得盔型溞的休眠种群可以在短时间内进入现生种群.盔型溞现生种群在经历了整个生长季节的克隆侵蚀后,在生长季节末期,盔型溞又通过有性生殖产生休眠卵,进入底泥,从而形成现生种群到休眠种群的基因流.可见,流溪河盔型溞现生种群与休眠种群始终存在双向的基因流,因此,休眠种群与现生种群没有出现较大的遗传分化,现生种群遗传多样性来自于休眠种群,因此,在水库中,休眠种群更能反映种群真实的遗传多样性.
| [1] |
Jiang Xiezhi, Du Nanshan. Fauna sinica; Crustacea: freshwater Cladocera. Beijing: Science Press, 1979. [蒋燮治, 堵南山. 中国动物志, 节肢动物门, 甲壳纲, 淡水枝角类. 北京: 科学出版社, 1979.]
|
| [2] |
Brendonck L, De Meester L. Egg banks in freshwater zooplankton: evolutionary and ecological archives in the sediment. Hydrobiologia, 2003, 491: 65-84. DOI:10.1023/A:1024454905119 |
| [3] |
Chesson PL, Warner RR. Environmental variability promotes coexistence in lottery competitive systems. American Naturalist, 1981, 117(6): 923-943. DOI:10.1086/283778 |
| [4] |
Ellner S, Hairston Jr NG. Role of overlapping generations in maintaining genetic variation in a fluctuating environment. American Naturalist, 1994, 143(3): 403-417. DOI:10.1086/285610 |
| [5] |
Caceres CE. Interspecific variation in the abundance, production, and emergence of Daphnia diapausing eggs. Ecology, 1998, 79: 1699-1710. DOI:10.1890/0012-9658(1998)079[1699:IVITAP]2.0.CO;2 |
| [6] |
Cousyn C, De Meester L, Colbourne JK et al. Rapid, local adaptation of zooplankton behavior to changes in predation pressure in the absence of neutral genetic changes. Proceedings of the National Academy of Sciences, 2001, 98(11): 6256-6260. DOI:10.1073/pnas.111606798 |
| [7] |
Brede N, Sandrock C, Straile D et al. The impact of human-made ecological changes on the genetic architecture of Daphnia species. Proceedings of the National Academy of Sciences, 2009, 106(12): 4758-4763. DOI:10.1073/pnas.0807187106 |
| [8] |
Boileau MG, Hebert PDN, Schwartz SS. Non-equilibrium gene frequency divergence: persistent founder effects in natural populations. Journal of Evolutionary Biology, 1992, 5: 25-39. DOI:10.1046/j.1420-9101.1992.5010025.x |
| [9] |
De Meester L, Vanoverbeke, De Gelas K et al. Genetic structure of parthenogenetic zooplankton populations-a conceptual framework. Archiv für Hydrobiologie, 2006, 167(4): 217-244. |
| [10] |
Spark P. Temporal changes in the genetic structure of the Daphnia species complex in Tjeukemeer, with evidence for backcrossing. Heredity, 1996, 76: 539-548. DOI:10.1038/hdy.1996.77 |
| [11] |
Spaak P, Denk A, Boersma M et al. Spatial and temporal patterns of sexual reproduction in a hybrid Daphnia species complex. Journal of Plankton Research, 2004, 26: 625-635. DOI:10.1093/plankt/fbh064 |
| [12] |
Xiang XL, Xi YL, Wen XL et al. Genetic differentiation and phylogeographical structure of the Brachionuscalyciflorus complex in eastern China. Molecular Ecology, 2011, 10: 3027-3044. |
| [13] |
Xiang Xianling, Tao Yuanyuan, Xi Yilong et al. Review on phylogeographical pattern and population genetic structure of rotifer. Chinese Journal of Ecology, 2015, 34(7): 2048-2057. [项贤领, 陶媛媛, 席贻龙. 轮虫种群遗传结构及系统地理格局研究进展. 生态学杂志, 2015, 34(7): 2048-2057.] |
| [14] |
Han Xiaoyu, Xu Lei, Chen Xiang et al. Diversity and spatial distribution of cladoceran resting eggs in surface sediments of Liuxihe Reservoir, Guangdong. Ecological Science, 2011, 30(3): 273-279. [韩小玉, 徐磊, 陈向等. 广东流溪河水库表层沉积物中枝角类休眠卵的多样性及空间分布. 生态科学, 2011, 30(3): 272-278.] |
| [15] |
Lin Qiuqi. Species diversity and spatial and temporal variations of pelagic metazooplankton in Liuxihe Reservoir [Dissertation]. Guangzhou: Jinan University, 2007. [林秋奇. 流溪河水库后生浮游动物多样性与群落结构的时空异质性[学位论文]. 广州: 暨南大学, 2007.]
|
| [16] |
Wang Tian. Predation and competition: Interspecies interactions of zooplankton communities in Liuxihe Reservoir [Dissertation]. Guangzhou: Jinan University, 2009. [望甜. 捕食与竞争——一座亚热带水库的浮游动物群落的种间关系研究[学位论文]. 广州: 暨南大学, 2009.]
|
| [17] |
Hairston NG, Brunt RAV, Kearns CM et al. Age and survivorship of diapausing eggs in a sediment egg bank. Ecology, 1995, 76(6): 1706-1711. DOI:10.2307/1940704 |
| [18] |
Limburg PA, Weider LJ. Ancient' DNA in the resting egg bank of a microcrustacean can serve as a palaeolimnological database. Proceedings of the Royal Society of London B, 2002, 269: 281-287. DOI:10.1098/rspb.2001.1868 |
| [19] |
Vandekerkhove J, Declerck S, Vanhove M et al. Use of ephippial morphology to assess richness of anomopods: potentials and pitfalls. Journal of Limnology, 2004, 63(1s): 75-84. DOI:10.4081/jlimnol.2004.s1.75 |
| [20] |
Montero-Pau J, Muñoz J. Application of an inexpensive and high-throughput genomic DNA extraction method for the molecular ecology of zooplanktonic diapausing eggs. Limnology and Oceanography Methods, 2008, 6(6): 218-222. DOI:10.4319/lom.2008.6.218 |
| [21] |
Xu Lei, Li Sijia, Wang Sheng et al. Comparison of two methods for extracting DNA of resting eggs by Cladocera. J Lake Sci, 2014, 26(4): 632-636. [徐磊, 李思嘉, 王晟等. 两种提取枝角类休眠卵基因组DNA方法的比较. 湖泊科学, 2014, 26(4): 632-636. DOI:10.18307/2014.0419] |
| [22] |
Prosser S, Martínez-Arce A, Elías-Gutiérrez M. A new set of primers for COI amplification from freshwater microcrustaceans. Molecular Ecology Resources, 2013, 13(6): 1151-1155. |
| [23] |
Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series, 1999, 41: 95-98. |
| [24] |
Rozas J, Sanchez-Delbarria JC, Messeguer X. DnaSP, DNA polymorphism analyses by the coalescent and othermethods. Bioinformatics, 2003, 19: 2496-2497. DOI:10.1093/bioinformatics/btg359 |
| [25] |
Kumar S, Nei M, Dudley J et al. MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Briefings in Bioinformatics, 2008, 9(4): 299-306. DOI:10.1093/bib/bbn017 |
| [26] |
Salzburger W, Ewing GB, Haeseler VA. The performance of phylogenetic algorithms in estimating haplotype genealogies with migration. Molecular Ecology, 2011, 20: 1952-1963. DOI:10.1111/mec.2011.20.issue-9 |
| [27] |
Xu S, Schaack S, Seyfert A et al. High mutation rates in the mitochondrial genomes of Daphnia pulex. Molecular Biology and Evolution, 2012, 29: 763-769. DOI:10.1093/molbev/msr243 |
| [28] |
Posada D, Crandall KA. Selecting the best-fit model of nucleotide substitution. Systematic Biology, 2001, 50: 580-601. DOI:10.1080/10635150118469 |
| [29] |
Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogeny. Bioinformatics, 2001, 17: 754-755. DOI:10.1093/bioinformatics/17.8.754 |
| [30] |
Huelsenbeck JP, Ronquist F, Nielsen R et al. Bayesian inference of phylogeny and its impact on evolutionary biology. Science, 2001, 294: 2310-2314. DOI:10.1126/science.1065889 |
| [31] |
Page RDM. TREEVIEW: an application to display phylogenetic trees on personal computers. Computer Applications in the Biosciences, 1996, 12: 357-358. |
| [32] |
De Meester L, Gomez A, Okamura B et al. The monopolization hypothesis and the dispersal-gene flow paradox in aquatic organisms. Acta Oecologica, 2002, 23: 121-135. DOI:10.1016/S1146-609X(02)01145-1 |
| [33] |
Rellstab C, Maurer V, Zeh M et al. Temporary collapse of the Daphnia population in turbid and ultra-oligotrophic Lake Brienz. Aquatic Sciences, 2007, 69(2): 257-270. DOI:10.1007/s00027-007-0872-7 |
| [34] |
Vanickova I, Jaromir SEDA, Machacek J et al. Effects of extreme floods on the Daphnia ephippial egg bank in a long narrow reservoir. Journal of Limnology, 2011, 70(2): 369-377. DOI:10.4081/jlimnol.2011.369 |
| [35] |
Snell TW. Sex, population dynamics and resting egg production in rotifers. Hydrobiologia, 1987, 144(2): 105-111. DOI:10.1007/BF00014523 |
| [36] |
Schwartz SS, Hebert PDN. Methods for the activation of the resting eggs of Daphnia. Freshwater Biology, 1987, 17(2): 373-379. DOI:10.1111/fwb.1987.17.issue-2 |
| [37] |
De Meester LD, De Jager H. Hatching of daphnia sexual eggs. 1. intraspecific differences in the hatching responses of daphnia-magna eggs. Freshwater Biology, 1993, 30(2): 219-226. DOI:10.1111/fwb.1993.30.issue-2 |
| [38] |
Vandekerkhove J, Declerck S, Jeppesen E et al. Dormant propagule banks integrate spatio-temporal heterogeneity in cladoceran communities. Oecologia, 2005, 142(1): 109-116. DOI:10.1007/s00442-004-1711-3 |
| [39] |
Vandekerkhove J, Declerck S, Brendonck L et al. Uncovering hidden species: hatching diapausing eggs for the analysis of cladoceran species richness. Limnology and Oceanography: Methods, 2005, 3(9): 399-407. DOI:10.4319/lom.2005.3.399 |
| [40] |
Vandekerkhove J, Declerck S, Brendonck L et al. Hatching of cladoceran resting eggs: temperature and photoperiod. Freshwater Biology, 2005, 50(1): 96-104. DOI:10.1111/fwb.2005.50.issue-1 |
| [41] |
Pérez-Martínez C, Jiménez L, Moreno E et al. Emergence pattern and hatching cues of Daphnia pulicaria (Crustacea, Cladocera) in an alpine lake. Hydrobiologia, 2013, 707(1): 47-57. DOI:10.1007/s10750-012-1404-6 |
 2017, Vol. 29
2017, Vol. 29 

